Abstract
Strong expression of MYC and MYC-regulated genes defines the molecular high-grade / double-hit signature subgroup of germinal center B-cell diffuse large B-cell lymphoma (GCB-DLBCL) which shows poor clinical outcomes with current therapy. About half of such cases show large-scale chromosomal rearrangements between the MYC locus and heterologous enhancer-bearing loci, but there is little understanding of the specific regulatory elements that activate MYC in absence of a rearrangement, or in the setting of non-immunoglobulin MYC rearrangement partners (RPs).
We used high-throughput CRISPR-interference functional profiling (CRISPRi) to pinpoint cis-regulatory element dependencies in MYC-intact and MYC-rearranged lymphoma cell lines at the level of individual nucleosome-free regions (NFRs). Custom sgRNA libraries targeted 233 individual NFRs (median 42 sgRNA / NFR after stringent quality filtering) across the MYC topological domain (TAD) and MYC RP loci in six GCB-DLBCL cell lines and two comparator mantle cell lymphoma (MCL) cell lines. Promoter-targeting sgRNAs against 587 sequence-specific transcription factor (TF) genes and 65 essential gene controls allowed for interrogation of trans-factor dependencies in the same assay.
CRISPRi profiling in one GCB-DLBCL and one MCL cell line with intra-chromosomal rearrangements of chromosome 8 showed strong, unique dependencies on enhancers within the cell-line-specific RPs, supporting an "enhancer-hijacking" function of these rearrangements.
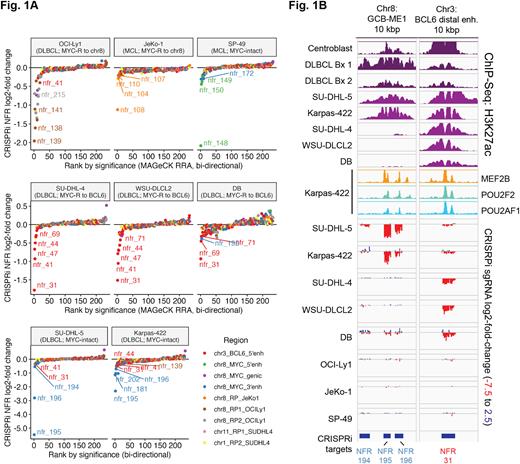
Three profiled cell lines showed structurally distinct variants of a recurrent t(3;8)(q27;q24) rearrangement that links the MYC gene to the distal super-enhancer region 150-240 kB upstream of BCL6. Although the distal BCL6 super-enhancers have been shown to sustain the essential BCL6 oncogene itself, we surprisingly found only weak dependency on the distal BCL6 enhancers for the three GCB-DLBCL cell lines lacking t(3;8) rearrangements. In contrast, all three GCB-DLBCL cell lines with t(3;8) rearrangements showed strong dependency on specific NFRs within the distal BCL6 super-enhancer. Validation assays showed that concurrent targeting of two BCL6 enhancer NFRs lead to even greater growth suppression and loss of MYC expression in t(3;8)-bearing cell lines.
In both GCB-DLBCL cell lines lacking MYC rearrangements, we identified a strong, novel dependency on a specific enhancer ("GCB-ME1") in the 3' portion of the MYC TAD. This enhancer was acetylated in MYC-intact GCB-DLBCL patient samples and normal centroblasts, but not in MCL or MYC-rearranged DLBCL samples. We confirmed that sustained MYC expression in MYC-intact cell lines requires activity of GCB-ME1. GCB-ME1 showed little or no essentiality in the four MYC-rearranged DLBCL cell lines or in the MYC-intact MCL cell line SP-49, which is dependent on previously identified Notch/RBPJ-regulated 5' MYC enhancers (Ryan et al., 2017).
ChIP-Seq showed that both GCB-ME1 and all of the most strongly essential BCL6 enhancers in t(3;8) cell lines were strongly bound by a triad of TFs, MEF2B, POU2F2, and POU2AF1, that were previously implicated in BCL6 activation. These genes were required to sustain MYC expression in both MYC-intact and t(3;8) GCB-DLBCL, and were highly essential in those cell lines, but POU2F2 and MEF2B were not essential in some cell lines with alternate MYC rearrangements.
Our data show that DLBCL is highly sensitive to selective disruption of discrete MYC-activating enhancers. Transcriptional regulatory factor dependencies in DLBCL may be shaped by the requirement to sustain MYC activation via the native GCB-ME1 enhancer or alternately via enhancers in specific RP loci, a finding with implications for therapeutic targeting of transcriptional regulatory mechanisms in DLBCL.
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal